Understanding your Genetic Test Result – Amino Acid Mutations
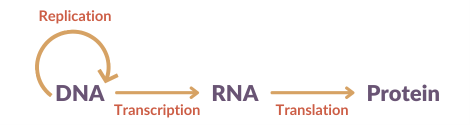
DNA is a unique code that contains all the instructions required for making the proteins needed for the development, growth and function of our bodies. The building blocks of our DNA are called nucleotides. There are four of these made of the bases: adenine, cytosine, guanine and thymine. The sequence of these determines our unique genetic code and tells the machinery in our cells what proteins to make, through an intermediate molecule called RNA. RNA carries the message of the DNA and allows it to be translated into proteins. It is very similar to DNA, however a base called uracil, replaces the base thymine.
Like DNA and RNA, proteins are also made of building blocks. These are not like those in DNA and RNA and are called amino acids. Our DNA is read in triplets: 3 bases (also called a codon) code for 1 amino acid. There are 20 different amino acids that can be made from the following different combinations:
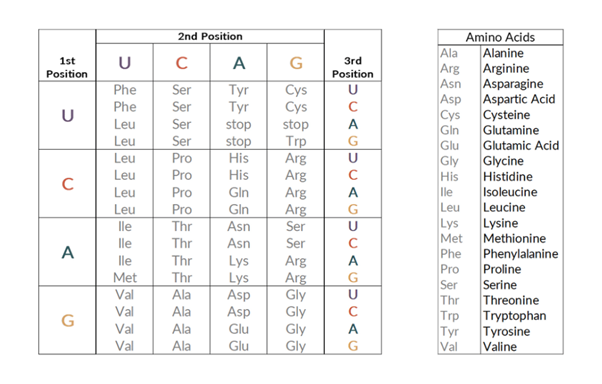 There are also 3 codons that do not code for an amino acid and instead signify for the protein to stop being made.
There are also 3 codons that do not code for an amino acid and instead signify for the protein to stop being made.
Interpreting Protein Mutations
Interpreting your protein mutation is very similar to interpreting your DNA mutation – you just need to know how to break it down into pieces!
Firstly the p. at the beginning tells us that this result is at the protein level.
The next three letters refer to the amino acid that is normally there in a non-mutated version of folliculin (see the above image).
The numbers that follow this refer to the position of the amino acid in the sequence.
The next bit(s) tell us what has happened to that particular amino acid at the position, for which there are a few different options
- The numbers may be followed by ‘del’, which doesn’t stand for an amino acid and means deletion.
e.g. p.(Phe157del) – the amino acid phenylalanine at position 157 has been deleted. - It is also possible for there to be a larger deletion.
e.g. p.(His111_Gln116del) – the amino acids from position 111 through 116 have been deleted. - There may also be another three letters referring to a different amino acid.
e.g. p.(Glu434Lys) – a Glutamine at position 434 has been swapped to a Lysine. - There also may be no letters after the numbers and there is only an *.
e.g. p.(Trp511*) – the amino acid tryptophan at position 511 has been mutated to a stop codon and the rest of the protein will not be made.
It is also possible that the result will look even more complicated as suggested above such as p.(His429Profs*27). Based on the above information it can be inferred that the amino acid histidine (His) at position 429 has been changed to a proline (Pro). However, the remaining fs*27 is yet to be explained.
The fs stands for frameshift. As our DNA is read in triplets, if a single amino acid is deleted or inserted it knocks the reading of triplets out of frame and means that the triplets are not read as they were designed. This can result in the introduction of a premature stop codon, as is the case with the p.(His429Profs*27). The fs*27 indicates that a frameshift has introduced a stop codon (*) 27 amino acids after the mutation.

It is also possible that your sequencing result may look like p.? – in which case the change at the protein level is unknown and cannot be predicted. This occurs when the type of DNA mutation is a splice-site mutation.
We hope you enjoy these toolkit posts and find them useful. If there is any aspect of BHD you would like us to do a deep dive on and explain in more detail, please let us know.
Previous
Back to